Results:
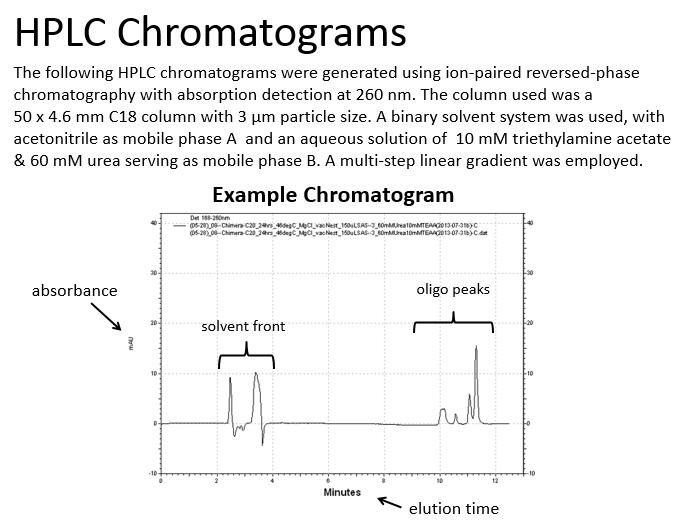
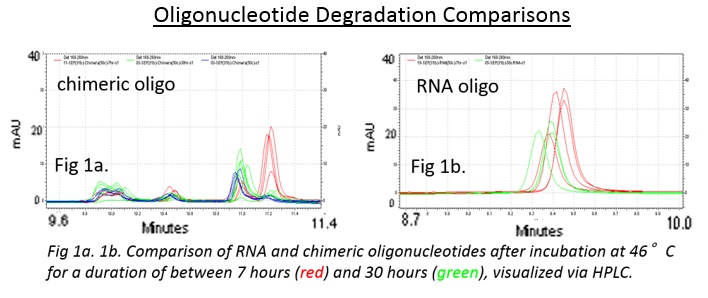
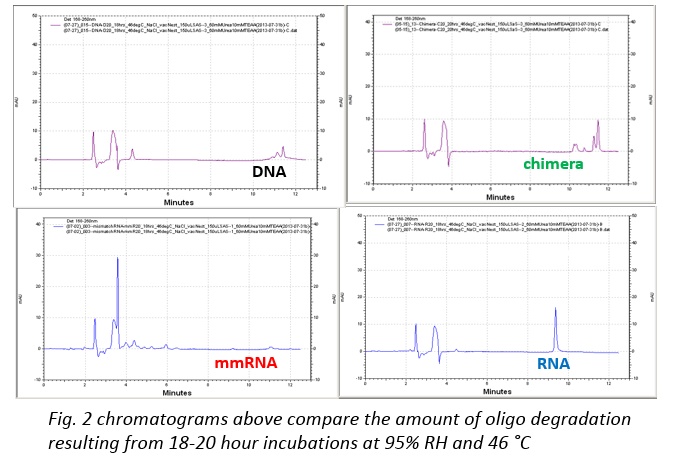
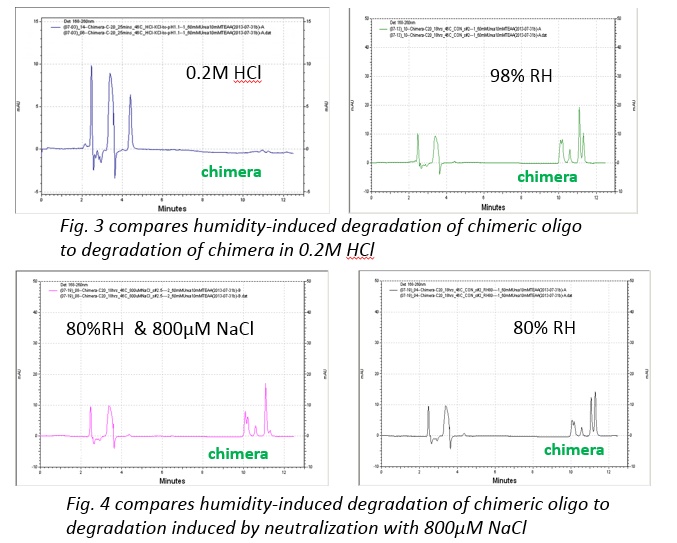
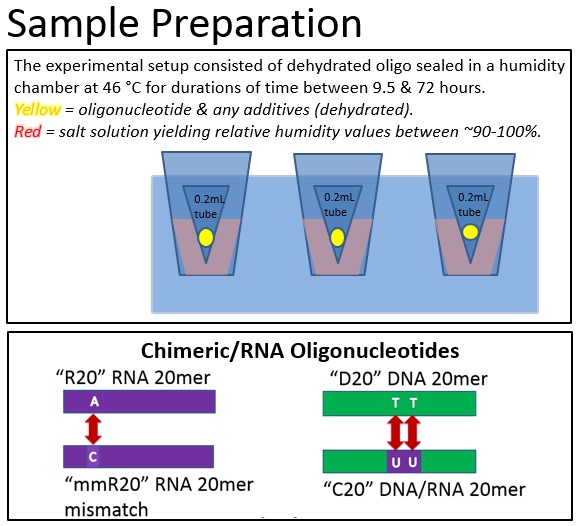
Conclusion:
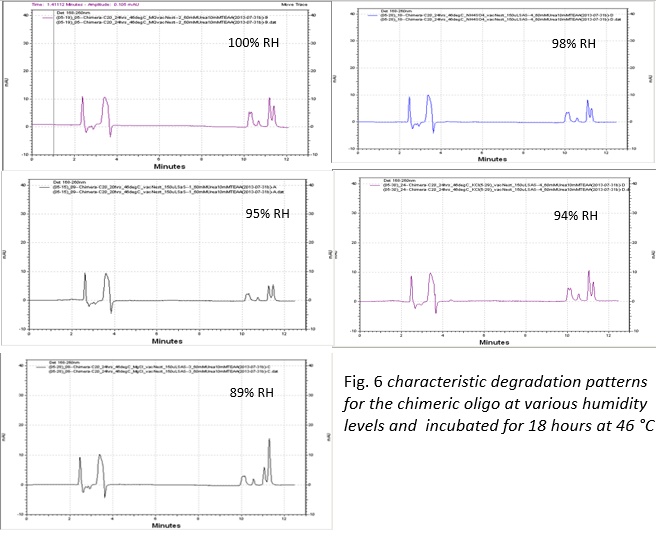
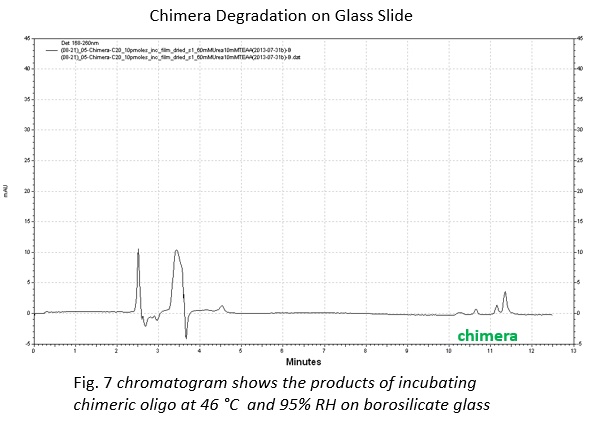
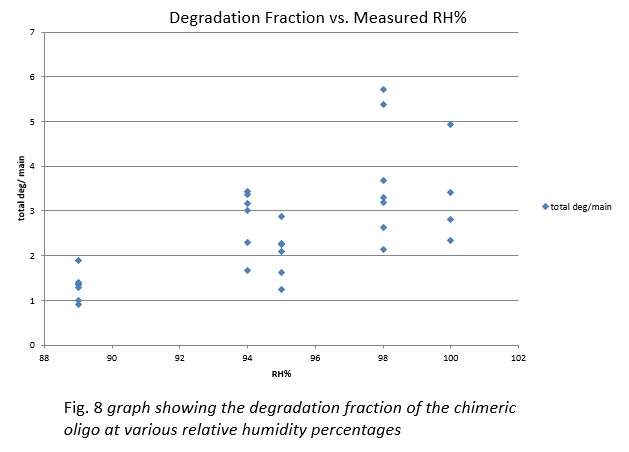
From this investigation, we were able to degrade chimeric oligonucleotides on multiple surfaces: polypropylene and borosilicate glass. Additionally, we found that the degradation on polypropylene can be controlled by modifying the humidity of the environment with saturated salt solutions.
Discussion:
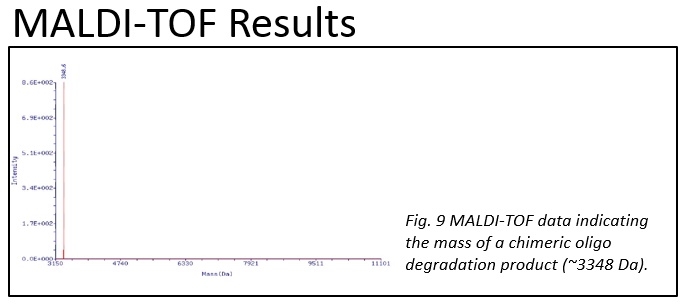
While we did find reliable and humidity-dependent degradation, the key issue that we face is that the mechanism of the reaction is still in debate. Understanding more about the mechanisms that drive this reaction could help us to optimize the process and better understand the implications and applications that may arise in the future. At first, the most likely driver of the degradation would seem to be hydrolysis of the phosphodiester linkages [1,2,3]. However, when this mechanism was tested by incubating the chimera at 46 °C in acidic solutions between pH 1-4.5 we did not observe a similar degradation pattern. Rather than producing products similar to those characteristic of humid degradation we observed a very different pattern. Thus suggesting that direct hydrolysis of the chimeric stand may not be the mechanism of reaction. Excluding non-neutral pH changes and addition of enzymes, there are still a number of known reaction that similarly produce ssDNA cleavage in the absence of acid, bases, or enzymes [4,5].
Another interesting aspect of this setting is the adsorption of the oligonucleotide to the surface and how this could be influenced by the humidity of the environment. Single-Stranded nucleotides, due to their more flexible structure, are able to more strongly absorb to negatively charged surfaces than double stranded oligos due to the difference in electrostatic interaction [6]. In this experiment, the environment around the oligos is very humid and likely influences the degree to which the oligos are adsorbed to the surfaces and this could be linked to the observations of different degradation at different humidity percentages. Additionally, electrostatic forces also play a key role in the adsorption of the oligonucleotide on a surface [7]. We have observed some cases where adsorption could be impacting the reaction but the implications of those results are still in debate. With regards to the sample recovery after incubation, we have seen that, in certain cases, adsorption can be so strong that the standard procedure of rehydration and pipette-mixing is unable to desorb any of the oligo from the tube. Because of the strength of the adsorption in these cases, no sample was able to be recovered which leaves the nature of the product in doubt.
It is also important to note that the degradation products on different surfaces do not appear consistent, while we were able to observe similar degradation patterns between surfaces the amount of degradation product was lower when compared to the original studies with the polypropylene tubes. Interestingly, this observation appears to have connections to both the mechanisms driving the reaction as well as the degree of adsorption of the oligos on the surfaces tested. That the oligos, when placed in a slightly different position in the humidity chamber, tend to exhibit lower degradation could help to illuminate some of the key conditions for this reaction to take place. As for the adsorption that oligos have to the surfaces tested, we found that in several cases after incubation no material was recovered from the surface. It is possible, that this is due to an error in the methodology but seems more likely this is driven by interactions with the surface that could make adsorption more likely and more effective [6]. This observation too appears to highlight an important characteristic of the reaction that may, as well, be an important factor governing the degradation of the chimeric oligonucleotides. Indeed, we have seen that when the oligos are incubated in a PCR-cycler, which differs from the heating block in that the solution cannot condense at the top of the tube, degradation was never observed. These results seem to indicate that strong adsorption to the tube could prevent degradation from taking place. While there is literature to suggest that in these low-humidity environments oligonucleotides do not behave in a typical manner [8,9], it is unknown just how changes in conformation could create the results observed.
[1] A. Blasko & T. Bruce “Recent Studies of Nucleophilic, General-Acid, and Metal Ion Catalysis of Phosphate Diester Hydrolysis” Accounts of Chemical Research. Vol. 32 NO. 6, 1999.
[2] Tomas Lindahl “Instability and decay of he primary structure of DNA” Nature Reviews Vol.362.1993.
[3] M. Oivanen et al. “Kinetics and Mechanisms for the Cleavage of Isomerization of the Phosphodiester Bonds of RNA by Bronstead Acids and Bases.” Chemical Reviews. Vol. 98, No. 3, 1998.
[4] R. Levis & L. Romano “Laser Vaporization of Single-Stranded DNA. A Study of Photoinduced Phosphodiester Bond Scission.” Journal of the American Chemical Society. 113, 7802-7803. 1991.
[5] H. Elsner & E. Lindblad “Ultrasonic Degradation of DNA” DNA. Vol 8, No. 10. 1989.
[6] H. Li & L. Rothberg “Colorimetric detection of DNA sequences based on electrostatic interactions with unmodified gold nanoparticles”PNAS. Vol. 101, No. 39. 2004.
[7] V.Dugas et al. “Immobilization of single-stranded DNA fragments to solid surfaces and their repeatable specific hybridization: covalent binding or adsorbtion?” Sensors and Actuators B. 101, 112-121. 2004.
[8] A. Montanari & M. Mezard “Hairpin formation and Elongation of Biomolecules” Physical Review Letters. Vol. 86, No. 10, 2178-2181. 2001.
[9] Goddard et al. “Sequence Dependent Rigidity of Single Stranded DNA” Physical Review Letters. Vol. 85, No. 11. 2400-2403. 2000.